Chocó, Colombia: a hotspot of human biodiversity
El Chocó, Colombia: un hotspot de la biodiversidad humana
Miguel A. Medina-Rivas1,2, Emily T. Norris3, Lavanya Rishishwar3,4, Andrew B. Conley3, Camila Medrano-Trochez5, Augusto Valderrama-Aguirre2,5, Fredrik O. Vannberg4, Leonardo Mariño-Ramírez2,6,7, I. King Jordan2,4,7
1 Grupo de Investigación en Biotecnología y Recursos
Fitogenéticos. Centro de Investigaciones en Biodiversidad y Hábitat, Universidad Tecnológica del Chocó,
Quibdó, Chocó, Colombia. Proyecto
"Desarrollo de herramientas de gestión para el posicionamiento
de la biodiversidad como fuente de bienestar social y
ambiental en el Chocó, occidente colombiano".
2 PanAmerican Bioinformatics Institute, Cali, Valle del Cauca, Colombia.
3 IHRC-Georgia Tech Applied Bioinformatics Laboratory, Atlanta, Georgia, USA.
4 School of Biology, Georgia Institute of Technology, Atlanta, Georgia, USA. e-mail: king.jordan@biology.gatech.edu
5 Biomedical Research Institute, Universidad Libre, Cali, Valle del Cauca, Colombia.
6 National Center for Biotechnology Information, National Institutes of Health, Bethesda, Maryland, USA.
7 BIOS Centro de Bioinformática y Biología Computacional, Manizales, Caldas, Colombia.
Date
received: October 16, 2015
Date approval: January 20,
2016 Asociated Editor: Jimenez AM.
Abstract
Objective: Chocó
is a state located on the Pacific coast of Colombia that has a majority
Afro-Colombian population. The objective of this study was to
characterize the genetic ancestry, admixture and diversity of the
population of Chocó, Colombia. Methodology:
Genetic variation was characterized for a sample of 101 donors (61
female and 40 male) from the state of Chocó. Genotypes were
determined for each individual via the characterization of 610,545
single nucleotide polymorphisms genome-wide. Haplotypes for the
uniparental mitochondrial DNA (female) and Y-DNA (male) chromosomes
were also determined. These data were used for comparative analyses
with a number of worldwide populations, including putative ancestral
populations from Africa, the Americas and Europe, along with several
admixed American populations. Results: The
population of Chocó has predominantly African genetic ancestry
(75.8%) with approximately equal parts European (13.4%) and Native
American (11.1%) ancestry. Chocó shows relatively high levels of
three-way genetic admixture, and far higher levels of Native American
ancestry, compared to other New World African populations from the
Caribbean and the United States. There is a striking pattern of
sex-specific ancestry in Chocó, with Native American admixture
along the female lineage and European admixture along the male lineage.
The population of Chocó is also characterized by relatively high
levels of overall genetic diversity compared to both putative ancestral
populations and other admixed American populations. Conclusion: These
results suggest a unique genetic heritage for the population of
Chocó and underscore the profound human genetic diversity that
can be found in the region.
Keywords: Admixture, Afro-Colombian, Colombia, Genetic ancestry, Genetic diversity, Human genome.
Resumen
Objetivo: El
Chocó es un departamento situado en la costa pacífica
colombiana cuya población es mayoritariamente afrocolombiana. El
objetivo de este estudio fue caracterizar la ancestralidad, el
mestizaje y la diversidad genética de la población del
Chocó colombiano. Metodología: La
variación genética se caracterizó en una muestra
de 101 donantes (61 mujeres y 40 hombres) del departamento del
Chocó. Los genotipos se determinaron para cada individuo a
través de la caracterización de 610,545 polimorfismos de
nucleótido único distribuidos en todo el genoma.
También se determinaron los haplotipos de los ADNs
uniparentales: ADN mitocondrial (materno) y cromosoma Y (paterno).
Estos datos se utilizaron para análisis comparativos en una
serie de poblaciones de todo el mundo, incluyendo poblaciones
ancestrales putativas de África, América y Europa,
además de varias poblaciones mestizas americanas. Resultados:
La población del Chocó tiene una ancestralidad
genética predominantemente africana (75,8%), con aportes
similares de ancestralidad europea (13,4%) y de nativos americanos
(11,1%). La población analizada del Chocó tiene niveles
relativamente altos de mestizaje triple y niveles mucho más
altos de ancestralidad nativa americana, en comparación con
otras poblaciones afrodescendientes del nuevo mundo, localizadas en
el Caribe y los Estados Unidos. Hay un patrón fuerte de
ancestralidad sexo-específica en el Chocó, con un
mestizaje predominantemente nativo americano en el linaje femenino y
europeo en el linaje masculino. La población del Chocó
también se caracteriza por niveles relativamente altos de
diversidad genética global en comparación con las
poblaciones ancestrales putativas y otras poblaciones mestizas
americanas. Conclusión:
Estos resultados sugieren un patrimonio genético único
para la población del Chocó y subrayan la profunda
diversidad genética humana que se puede encontrar en la
región.
Palabras clave: Afrocolombianos, Ancestralidad genética, Colombia, Diversidad genética, Genoma humano, Mestizaje.
Introduction
Chocó, Colombia.
Chocó is a Colombian administrative department (i.e., a state)
located along the country’s Pacific coast (Figure 1A).
Chocó ranges from the Panamanian border in the north to the
Cauca Valley in the southwest region of Colombia. The state of
Chocó is recognized worldwide as a hotspot of biodiversity
(http://www.eoearth.org/view/article/150631/). A biodiversity hotspot
is defined as a specific geographic region with a large amount of
endemic biodiversity that is threatened by human activity (Zachos and
Habel 2011). In order for an area to be officially recognized as a
biodiversity hotspot, at least 0.5% of its vascular plant species (or
1,500 species) must be characterized as endemics, which are defined as
species found uniquely within a proscribed geographic region or habitat
type. There are 25 global regions that qualify as biodiversity hotspots
according to this criterion, and together these areas are home to
almost 60% of the world’s known species of plants, birds,
reptiles, amphibians and mammals. The entire area of Chocó is
contained within the so-called Chocó-Darién biodiversity
hotspot, also known as the Tumbes-Chocó-Magdalena biodiversity
hotspot
(http://tmalliance.org/about/where-we-work/biodiversity-hotspot/).
The
Chocó-Darién biodiversity hotspot extends along the
Pacific coast from the Panamá Canal region in the north, through
the Darién Gap and Chocó wet rain forests in
Panamá-Colombia, passing completely through Ecuador before
ending in the coastal dry forests of northern Perú. This area
encompasses a wide variety of diverse habitats including the wettest
rain forests in the world, which are found in Chocó. The
Colombian portion of this biodiversity hotspot in Chocó is
relatively preserved compared to the Ecuadorean zone where 98% of the
native forest has been cleared. The Chocó-Darién
biodiversity hotspot supports ~10,000 species of vascular plants along
with 600 species of birds, 235 species of mammals, 350 species of
amphibians and 210 species of reptiles.
Human biodiversity in Chocó. Although
Chocó is widely recognized as a hotspot of biodiversity, one
critical aspect of the region’s biodiversity remains largely
unexplored, the diversity of its human population (Figure 1B).
The population of Chocó has a uniquely African genetic heritage
with admixture from the Americas and Europe. The vast majority of the
population is Afro-Colombian (82.1%) but there are also substantial
numbers of Native Americans (12.7%) and individuals with primarily
European ancestry (5.2%) (Hernández Romero 2005). There are
anywhere from 9 to 20 million Afro-descendants in Colombia, making it
the country with the third most Afro-descendants in the Americas.
Despite the presence of such a large population of Afro-descendants in
Colombia, there is a vast under-representation of genetic studies of
Afro-Colombians (Rishishwar et al. 2015a). Previous Colombian genetic
ancestry studies have dealt mainly with Mestizo genomes that have
primarily European and secondary Native American ancestry
(Carvajal-Carmona et al. 2000, Carvajal-Carmona et al. 2003, Bedoya et
al. 2006, Wang et al. 2008, Bryc et al. 2010, Córdoba et al.
2012, Ruiz-Linares et al. 2014, Rishishwar et al. 2015b). Given the
high percentage of Afro-Colombians living in Chocó, genetic
studies of this population are ideally suited to uncover the as yet
untapped African dimension of Colombian ancestry and human
biodiversity.
Colombian individuals
have three-way genetic admixture patterns that result from ancestral
contributions to the modern population from Africa, the Americas and
Europe (Rishishwar et al. 2015b). The story of every Colombian’s
ancestry, along with their specific admixture patterns, is written in
the sequence of their genome. Thus, genome sequence analysis can be
used to infer genetic ancestry and admixture patterns for individuals
and for the population as a whole. For individuals, the total overall
proportions of African, Native American and European ancestry can be
inferred along with both locus-specific and sex-specific patterns of
ancestry and admixture. Genetic ancestry can be explored at both the
continental level to uncover the broad regions of origin for Colombians
and at the sub-continental level to explore the specific ancestral
regions and countries from which individuals’ ancestors
originate. At the population level, locus-specific admixture patterns
can reveal whether natural selection has enriched for specific ancestry
along particular genomic segments. Population level inference can also
be used to assess whether there are sex-specific differences in
ancestry that result from differential ancestry contributions along
maternal versus paternal lineages. These kinds of genomic research
approaches will be applied in order to elucidate the patterns of
genetic ancestry and admixture of the population of Chocó under
the auspices of a newly formed collaborative research project ChocoGen
(http://www.chocogen.com/).
ChocoGen project. An
exploration of human genetic biodiversity in Chocó is being
conducted via the collaborative ChocoGen research project in an effort
to value, conserve and utilize this precious resource. The ChocoGen
research project has two overarching goals:
1) to characterize the genetic diversity and ancestry of the population of Chocó, and
2) to create a health profile of the region based on the genetic diversity of its people.
Research and development
activities in support of both of these goals are being conducted in
such a way as to develop the local human capacity in Chocó for
research and education in genetic health and medicine. This project is
a collaboration between Universidad Tecnológica del Chocó
(UTCH) in Colombia, principle investigator Dr. Miguel A. Medina Rivas,
and the Georgia Institute of Technology in the USA, principle
investigator Dr. I. King Jordan. Bioinformatics analysis and
interpretation of human genome sequences from the population of
Chocó are being further supported by the National Center for
Biotechnology Information (NCBI) in the USA, and the Colombian National
Center for Bioinformatics and Computational Biology (BIOS) in
Manizales.
Researchers from the
ChocoGen project are conducting analysis of genomic sequences sampled
from volunteers from the population of Chocó to characterize:
1) their genetic ancestry,
2) the quantity and nature of genetic admixture between ancestral populations, and
3) the possible relationship between genetic ancestry, admixture and determinants of health and disease.
The results of this project
will serve as a resource for the development and application of genetic
approaches to healthcare in the Pacific region of Colombia and help to
position UTCH as a leader in this area of applied research. In this
report, we present results of the first round of analyses of the
genetic ancestry of 101 individuals sampled from the population of
Chocó.
Methodology
Sample donors and genotyping. ChocoGen
volunteer DNA sample donors were recruited at UTCH. Donors were
selected in an effort to include representative samples of different
geographic regions of Chocó (Atrato, Baudó, Atlantic
coast, Pacific coast, San Juan), as well as an approximately equal
representation of males and females, and donors were asked to
self-identify their ethnic origins. Donors contributed DNA using a
non-invasive saliva sampling method. All donors signed informed consent
documents indicating their understanding of the potential risks of the
project along with how their data would be handled and how their
identity would be protected. Collection, genotyping and comparative
analyses of human DNA samples were conducted with the approval of the
ethics committee of UTCH. Donor DNA samples were genotyped using the
Illumina HumanOmniExpress-24 single nucleotide polymorphism (SNP) chip.
Comparative genomic data sources.
The genotypes of ChocoGen donors were compared to whole genome sequence
data from the 1000 Genomes Project (1000G) (Genomes Project et al.
2010, Genomes Project et al. 2015) and genotype data from the Human
Genome Diversity Project (HGDP) (Cann et al. 2002, Li et al. 2008) (Table 1).
Genotypes from the ChocoGen donors, along with genotypes from the 1000G
and HGDP projects, were all mapped to the coordinate space of the
February 2009 human genome reference sequence version GRCh37/hg19
(Lander et al. 2001, Kent et al. 2002) for subsequent analysis. The
program PLINK (Purcell et al. 2007) was used for genotype quality
control and to extract autosomal genotyped positions (i.e., single
nucleotide polymorphisms or SNPs) common to all three genotype sources
to yield a final merged genotype dataset. For quality control, only
individual SNP positions with a genotyping rate >98% were retained
for subsequent analysis.
Ancestry and admixture inference. The
program PLINK was used to prune the final merged genotype dataset by
removing correlated sets of SNPs. Genomic distances were computed as
pairwise allele sharing distances between all individual pruned
genotypes using PLINK. The resulting pairwise distance matrix was
projected onto two-dimensions with principal component analysis (PCA)
using the prcomp function from the R package for statistical computing
(Team 2008). The program ADMIXTURE (Alexander et al. 2009) was run on
the genotype dataset to infer individual ancestry components. The
resulting data was used with the nnls package from R to implement a
non-negative least squares method to estimate the fractions of African,
Native American and European ancestry for each individual from
Chocó. For each individual, the entropy (H) of the admixture was
calculated as

where
pi is the ancestry fraction for population i. Sex-specific ancestry in
the population of Chocó was determined via analysis of
uniparental haplotypes: mitochondrial DNA (mtDNA) for the maternal
lineage and Y chromosomal DNA (Y-DNA) for the paternal lineage.
The relative genetic
diversity levels of the populations analyzed here were measured via the
total amount of observed pairwise allele sharing distance and
genome-wide heterozygosity. The total amount of observed pairwise
allele sharing distance within each population was computed by fitting
a minimum spanning ellipse to the individual genotype points of the
population projected onto the first two principal components of the PCA
analysis using the ellipsoidhull function in R. The areas (A) of the
population-specific ellipses were computed using the lengths of the
major (x) and minor (y) axes scaled to the principal component weights:

Heterozygosity
was measured as the fraction of all genotype positions that are
heterozygous within an individual using the program PLINK. To do this,
we analyzed SNPs with minor allele frequency >25% in order allow for
comparison between SNPs called from genome sequences versus SNPs called
from genotype arrays, which are biased to high minor allele frequencies
and European populations.
Results and discussion
Genetic characterization of the population of Chocó. Volunteer
DNA sample donors were solicited on the main campus of UTCH located in
the capital city of Quibdó; 101 volunteers (61 females and 40
males) provided DNA samples for genetic characterization along with
answers to a series of questions related to their ethnic self-identity
and family history. DNA samples were characterized in order to
determine the specific identity of genetic sequence variants at 610,545
loci across the genome. Genetic variants are referred to here as single
nucleotide polymorphisms (SNPs), and the specific identity of the DNA
sequence residues that correspond to a genome-wide collection of SNPs
is referred to as a genotype. The specific identify of the DNA sequence
residues for a set of genetically linked SNPs is referred to as a
haplotype. For the purposes of this study, donors’ genotypes were
characterized for the entire set of human autosomes, and haplotypes
were determined for uniparental mitochondrial DNA (mtDNA) and Y-DNA
chromosomes. Chocó genotypes were compared to genotypes for a
variety of human populations (Table 1), in
order to make inferences about the genetic ancestry and diversity of
the population. Chocó mtDNA and Y-DNA haplotypes were compared
to known global distributions for haplotypes of these chromosomes in
order to make inferences about female-specific (mtDNA) and
male-specific (Y-DNA) genetic ancestry of the population.
Genetic ancestry and admixture of Chocó. The
Colombian population has a mixture of genetic ancestry from African,
European and Native American populations, owing to the historical
patterns of conquest and colonization in the New World (Markham 1912,
Mann 2013). Thus, Chocó genotypes were compared to genotypes
characterized from individuals sampled from representative populations
of these regions (Table 1) in order to infer
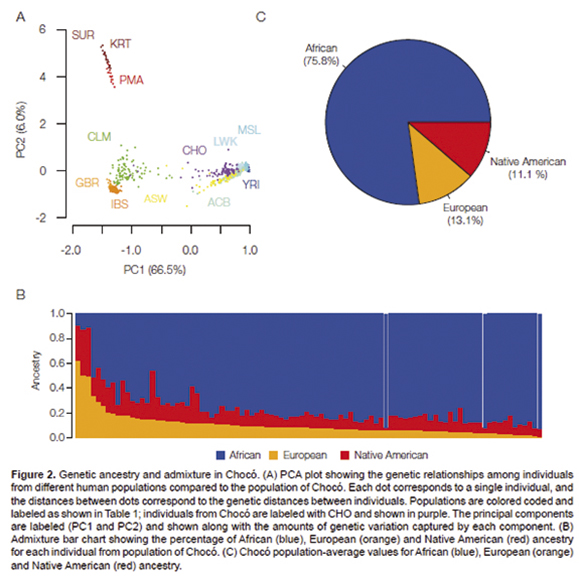
their overall genetic ancestry and admixture. The genetic relationships
among individuals from the population of Chocó, along with
individuals from the other global populations, are visually represented
in Figure 2A. This panel shows a
two-dimensional principal component analysis (PCA) projection of the
pairwise genetic distances between all of the genotypes analyzed here,
where the distance between each dot corresponds to the distance between
each individual genotype. The main component of human genetic diversity
in this representation is projected along the x-axis (PC1=66.5% of the
diversity) and the secondary component is shown on the y-axis (PC2=6%).
African, European and Native American populations occupy the three
poles of human genetic diversity in this plot, whereas admixed American
genomes, including Chocó (CHO) individuals, occupy intermediate
positions between these three ancestral, and relatively non-admixed,
population groups. The relative positions of the admixed American
populations compared to the three ancestral groups gives an indication
of their admixture proportions. For example, the Colombian population
from Medellín (CLM) shows evidence of more European admixture
compared to the Chocó population, which is located in much
closer proximity to the African populations. Two other New World
African populations (ACB and ASW) are also located in close proximity
to the putative ancestral populations from the African continent, but
occupy different positions than the Chocó population.
These same pairwise
genetic distances can be used to quantify the amount of genetic
ancestry that any admixed individual shows from the putative ancestral
African, European and Native American populations. The results of this
kind of analysis are shown in Figure 2B and 2C.
African ancestry represents the dominant admixture component for the
vast majority of individuals from the population of Chocó
analyzed here. The maximum fraction of African ancestry seen for any
individual is 92.8%, and the average African ancestry for Chocó
population is 75.8%. Nevertheless, there are substantial fractions of
European and Native American ancestry seen for many of these
individuals as well. The maximum fraction of European ancestry seen for
any individual is 62.8%, and the average European ancestry is 13.1%.
The maximum fraction of Native American ancestry is 39.6%, and the
average Native American ancestry is 11.1%. The broad range of
individual admixture percentages point to the diversity of the
Chocó population.
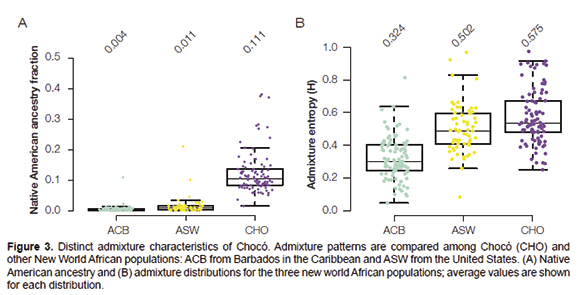
The genetic ancestry of
the population of Chocó shows some interesting differences
compared to the genetic ancestry of the two other New World African
populations analyzed here, the ACB population from Barbados in the
Caribbean and the ASW population from the USA, despite the fact that
all three populations show similarly high levels of overall African
ancestry (~75-80%). First of all, the Chocó population (CHO) has
substantially higher levels of Native American ancestry compared to the
Afro-Caribbean (ACB) or African-American (ASW) populations (Figure 3A).
Chocó has 11.1% average Native American ancestry, whereas the
Afro-Caribbean and African-American populations have 0.4% a 1.1%
average Native American ancestry, respectively. Second, the
Chocó population shows higher levels of three-way genetic
admixture, as measured by Admixture entropy (H), compared to the other
two New World African populations (Figure 3B).
This reflects the fact that in Chocó the non-African ancestry
component is relatively evenly divided between European and Native
American ancestry, whereas almost all non-African ancestry in the
Caribbean and US populations is European. This pattern is indicative of
longer and more sustained contact between Afro-descendants and
Indigenous communities in Chocó compared to what occurred in the
Caribbean or the United States.
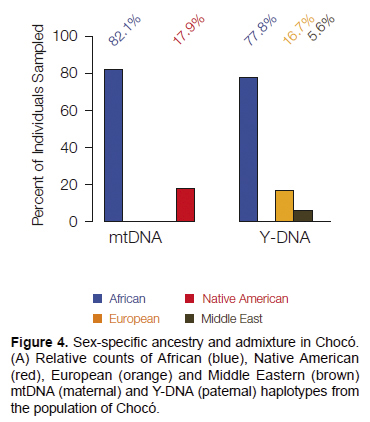
Sex-specific genetic ancestry. Mitochondrial
DNA (mtDNA) and Y-DNA chromosomes are referred to as uniparental
ancestry markers since they are inherited strictly along the maternal
(for mtDNA) and paternal (Y-DNA) lineages. This means that mtDNA
haplotypes can be used to infer female-specific ancestry and admixture,
and Y-DNA haplotypes can be used to infer male-specific ancestry and
admixture. The global origins of the mtDNA and Y-DNA haplotypes
characterized from the population of Chocó show striking
evidence of sex-specific ancestry in this population (Figure 4).
The majority of mtDNA (82.1%) and Y-DNA (77.8%) haplotypes have African
origins, consistent with the overall genetic ancestry of the
population. However, the non-African ancestry components differ
markedly for the female (mtDNA) versus male (Y-DNA) lineages. All of
the non-African mtDNA haplotypes (17.9%) have Native American origins,
whereas all of the non-African Y-DNA haplotypes have European (16.7%)
or Middle Eastern (5.6%) origins. This sex-specific pattern of genetic
ancestry may be linked to the unique historical conditions under which
the state of Chocó was founded and populated (Wade 1995).
Genetic diversity levels in Chocó.
As previously noted, based on its distinct population demographics, we
propose that the state of Chocó is a rich source of human
biodiversity. To evaluate this proposition with respect to the genetic
ancestry of the population, we compared the genetic diversity levels
found in Chocó to levels of diversity seen for putative
ancestral, non-admixed populations as well as other admixed American
populations. The results of this analysis are shown in Figure 5.
We evaluated genetic diversity in two ways: 1) via the overall scope of
genetic distances between individuals in a population and 2) via the
average genome-wide heterozygosity levels for all individuals in a
population. The overall genetic diversity for each population was
inferred by fitting a minimal spanning ellipse to the
populations’ pairwise genetic distance projection on the PCA plot
(Figure 5A). The areas of the population-specific minimum spanning
ellipses were then determined and used to quantify the
populations’ genetic diversity (Figure 5B). The Chocó
population (CHO) has the highest level of overall genetic diversity
calculated in this way for any of the populations analyzed here. The
Chocó population also has high average heterozygosity levels
compared to the other populations, second only to the other New World
African population ASW (Figure 5C). Interestingly, the other admixed
Colombian population from Medellín (CLM) also shows relatively
high genetic diversity levels in these analyses despite the fact that
it has a very different genetic ancestry profile (i.e., largely
European ancestry) compared to the population of Chocó.
Conclusions
The ChocoGen collaborative
research project has the joint aims of 1) characterizing the ancestry
and genetic diversity of the population of Chocó, and 2)
creating a genetic health profile of the population based on the
diversity of its people. Investigators from UTCH and the Georgia
Institute of Technology are collaborating to these ends, and this
manuscript reports some of the first results of the project. The
initial phase of the project is focused on ancestry analysis, and the
results from this first phase will be used to inform the second
health-related part of the effort. All of this work is being done in
such a way as to develop the local human capacity for research in
genetic ancestry and human health in the state of Chocó.
The analyses reported
here indicate that the population of Chocó has an overwhelmingly
African genetic ancestry, which is of course not surprising.
Nevertheless, the Chocó population shows interesting
differences, compared to other admixed American populations with
similar levels of African ancestry, which likely reflect its distinct
historical and cultural traditions (Wade 1995). In particular,
individuals from Chocó show higher levels of three-way genetic
admixture than other New World African populations, and this pattern
can be largely attributed to the higher levels of Native American
ancestry seen in Chocó. The population of Chocó also
shows striking patterns of sex-specific ancestry, whereby non-African
maternal ancestry is exclusively Native American, and non-African
paternal ancestry is almost entirely European. This ancestry pattern
may represent distinct admixture dynamics that characterized early
(European admixture) from later (Native American admixture) historical
periods in Chocó, and we plan to explore this idea further in
subsequent studies.
Finally, the results on
genetic ancestry and diversity obtained in this study underscore the
extent to which Chocó represents a hotspot of human
biodiversity. We hold that the human biodiversity of Chocó is an
under-appreciated dimension of the area’s well known
biodiversity, and one that should be equally valued and fully developed
for its potential.
Literature cited
- Alexander DH, Novembre J,
Lange K. 2009. Fast model-based estimation of ancestry in unrelated
individuals. Genome Res. 19 (9): 1655-64.
- Bedoya G, Montoya P,
García J, Soto I, Bourgeois S, Carvajal L, et al. 2006.
Admixture dynamics in Hispanics: a shift in the nuclear genetic
ancestry of a South American population isolate. Proc Natl Acad Sci
USA. 103 (19): 7234-9.
- Bryc K, Vélez C,
Karafet T, Moreno-Estrada A, Reynolds A, Auton A, et al. 2010.
Colloquium paper: genome-wide patterns of population structure and
admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA 107
(Suppl 2): 8954-61.
- Cann HM, de Toma C, Cazes
L, Legrand MF, Morel V, Piouffre L, et al. 2002. A human genome
diversity cell line panel. Science. 296 (5566): 261-2.
- Carvajal-Carmona LG,
Ophoff R, Service S, Hartiala J, Molina J, Leon P, et al. 2003. Genetic
demography of Antioquia (Colombia) and the Central Valley of Costa
Rica. Hum Genet. 112 (5-6): 534-41.
- Carvajal-Carmona LG, Soto
ID, Pineda N, Ortiz-Barrientos D, Duque C, Ospina-Duque J, et al. 2000.
Strong Amerind/white sex bias and a possible Sephardic contribution
among the founders of a population in northwest Colombia. Am J Hum
Genet. 67 (5): 1287-95.
- Córdoba L,
García J, Hoyos LS, Duque C, Rojas W, Caravajal S, et al. 2012.
Composicion genética de una población del suroccidente de
Colombia. Rev Colomb Antropol. 48 (1): 21-48.
- Genomes Project C,
Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. 2010. A
map of human genome variation from population-scale sequencing. Nature.
467 (7319): 1061-73.
- Genomes Project C, Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. 2015. A global
reference for human genetic variation. Nature. 526 (7571): 68-74.
- Hernández Romero
A. 2005. La visibilización estadística de los grupos
étnicos colombianos. Bogotá: Departamento Administrativo
Nacional de Estadística (DANE).
- Kent WJ, Sugnet CW, Furey
TS, Roskin KM, Pringle TH, Zahler AM, et al. 2002. The human genome
browser at UCSC. Genome Res. 12 (6): 996-1006.
- Lander ES, Linton LM,
Birren B, Nusbaum C, Zody MC, Baldwin J, et al. 2001. Initial
sequencing and analysis of the human genome. Nature. 409 (6822):
860-921.
- Li JZ, Absher DM, Tang H,
Southwick AM, Casto AM, Ramachandran S, et al. 2008. Worldwide human
relationships inferred from genome-wide patterns of variation. Science.
319 (5866): 1100-4.
- Mann CC. 2013. 1493: Uncovering the new world Columbus created. New York: Alfred A. Knopf.
- Markham C 1912. The conquest of New Granada. New York: EP Dutton and Company.
- Purcell S, Neale B,
Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. 2007. PLINK: a
tool set for whole-genome association and population-based linkage
analyses. Am J Hum Genet. 81 (3): 559-75.
- Rishishwar L, Conley AB,
Vidakovic B, Jordan IK. 2015a. A combined evidence Bayesian method for
human ancestry inference applied to Afro-Colombians. Gene. 574 (2):
345-51.
- Rishishwar L, Conley AB,
Wigington CH, Wang L, Valderrama-Aguirre A, Jordan IK. 2015b. Ancestry,
admixture and fitness in Colombian genomes. Sci Rep. 5: 12376.
- Ruiz-Linares A, Adhikari
K, Acuna-Alonzo V, Quinto-Sánchez M, Jaramillo C, Arias W, et
al. 2014. Admixture in Latin America: geographic structure, phenotypic
diversity and self-perception of ancestry based on 7,342 individuals.
PLoS Genet. 10 (9): e1004572.
- Team RDC. 2008. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
- Wade P. 1995. Blackness and race mixture: the dynamics of racial identity in Colombia. Balitmore: JHU Press.
- Wang S, Ray N, Rojas
W, Parra MV, Bedoya G, Gallo C, et al. 2008. Geographic patterns
of genome admixture in Latin American Mestizos. PLoS Genet 4 (3):
e1000037.
- Zachos FE, Habel JC.
2011. Biodiversity hotspots: distribution and protection of
conservation priority areas. Vienna: Springer Science & Business
Media.